基于高通量测序的植物抗逆研究方案 返回
基于高通量测序的植物抗逆研究方案
•植物抗逆相关项目经验
基迪奥在过去两年内已完成约200个植物抗逆抗病项目,项目金额占比达18.6%,涉及物种覆盖模式植物、作物、林木、中草药等,项目经验丰富,专业服务有保障!

•植物抗逆全方位研究策略
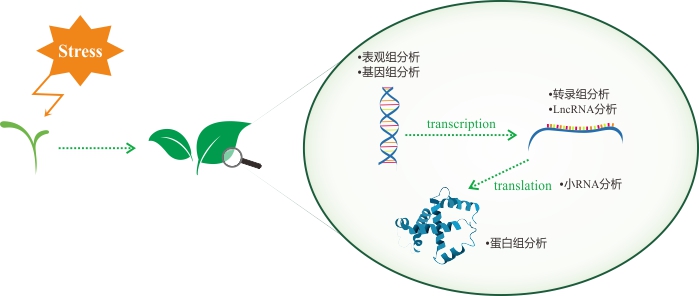
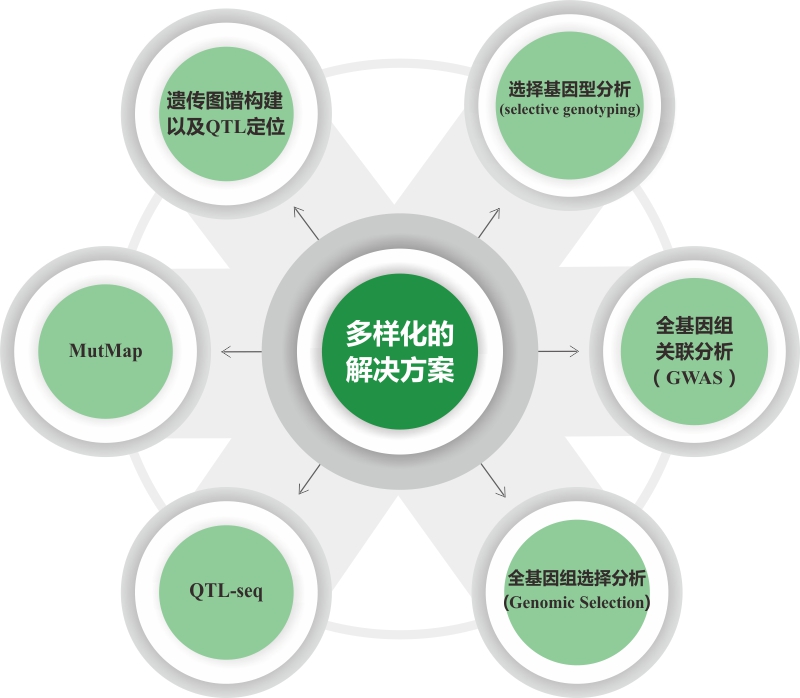
•基于基因组测序的植物抗逆基因定位解决方案
在经典遗传学理论中,DNA突变是性状差异的基础。因此,抗逆相关基因的定位是植物逆境胁迫研究的重要基础。我们会根据您的材料特性、研究目的和经费预算,提供定制化的基因定位解决方案,加速功能基因挖掘。
案例解读
利用MutMap技术快速培育出水稻耐盐新品种[1]
为应对日本2011年地震和海啸引起的大面积稻田被海水淹没而造成的盐污染问题,研究者利用MutMap技术,鉴定出与水稻耐盐性状相关的基因OsRR22,该基因中的一个无义突变导致了水稻产生耐盐性。将耐盐突变体hst1与野生型亲本Hitomebore杂交,在短短两年时间内培育出了耐盐品种“Kaijin”,该品种保持了亲本优良的产量性状和品质性状,同时具有耐盐性,并在2015年在当地推广。
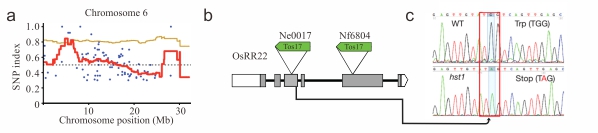
图1. 利用MutMap技术,鉴定出水稻6号染色体上OsRR22基因上的一个无义突变导致突变体hst1产生耐盐性
参考文献
[1]Takagi H, Tamiru M, Abe A, et al. MutMap accelerates breeding of a salt-tolerant rice cultivar[J]. Nature biotechnology, 2015, 33(5): 445-449.
•基于RNA测序和多组学贯穿分析的抗逆调控机制研究方案
除了经典遗传学外,植物在各种逆境胁迫下的基因调控机制,尤其是表观修饰调控变化,是今年来的研究热点。我们将根据您的材料特性、潜在的关键调控因子、期望目标等,提供定制化的不同组学调控机制解决方案,助您逐步深入挖掘逆境胁迫调控的分子机制。
1. 灵活的实验设计与方案
不同的样本数和实验处理有不同的解决方案:
对照组vs处理组:两两差异分析,找出逆境导致的差异
梯度样本:基因表达趋势分析、聚类分析,找出基因表达的时空规律
多组样本、大样本量:WGCNA基因网络分析,综合描述基因间的调控关系
2、多组学贯穿运用,全面解决生物学问题
基因表达谱-miRNA贯穿分析:揭示转录调控、转录后调控机制
转录组-蛋白组贯穿分析:揭示基因转录与表达间的关系与调控机制
转录组-表观调控贯穿分析:研究植物压力记忆形成机制
案例解读
干旱和盐胁迫抗性不同的水稻品系间的DNA甲基化调控差异[2]
为了阐明水稻在非生物胁迫下的DNA甲基化调控机制,对三种品系(分别为抗盐、抗旱和胁迫敏感的品系)的水稻种子分别进行甲基化、转录组和小RNA检测。结果发现与差异甲基化相关的差异表达基因,显著与非生物逆境胁迫相关。而且也发现差异甲基化的转座子对周边的蛋白编码基因的表达有所影响。另外,小RNA潜在通过RdDM(RNA介导的DNA甲基化)对DNA甲基有调控作用。这些结果从甲基化修饰、基因转录表达和小RNA调控三个层面,对水稻的非生物逆境胁迫机理进行了阐述。
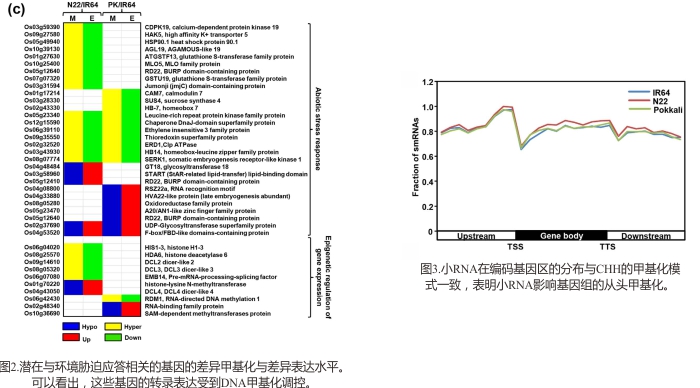
参考文献:
[2]Garg R, Chevala V V S N, Shankar R, et al. Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response[J]. Scientific reports, 2015, 5.
•部分已发表植物学相关文章
1.XU W, et al.(2016)Genomic DNA methylation analyses reveal the distinct profiles in castor bean seeds with persistent endosperms. Plant Physiology.
2.Wang W, Li R, Zhu Q, Tang X, & Zhao Q (2016) Transcriptomic and physiological analysis of common duckweed Lemna minor responses to NH4 + toxicity. BMC Plant Biology 16(1):1-13.
3.Zhang J, et al. (2016) Transcriptome Analysis of Dendrobium officinale and its Application to the Identification of Genes Associated with Polysaccharide Synthesis. Frontiers in plant science 7:5.
4.Sun Q, et al. (2016) To Be a Flower or Fruiting Branch: Insights Revealed by mRNA and Small RNA Transcriptomes from Different Cotton Developmental Stages. Scientific reports 6:23212.
5.Qu D, et al. (2016) Identification of MicroRNAs and Their Targets Associated with Fruit-Bagging and Subsequent Sunlight Re-exposure in the "Granny Smith" Apple Exocarp Using High-Throughput Sequencing. Frontiers in plant science 7:27.
6.Ma B, Li T, Xiang Z, & He N (2015) MnTEdb, a collective resource for mulberry transposable elements. Database : the journal of biological databases and curation 2015.
7.Liu H, et al. (2015) The Impact of Genetic Relationship and Linkage Disequilibrium on Genomic Selection. PloS one 10(7): e0132379.
8.Liu H, et al. (2015) An ultra-high-density map as a community resource for discerning the genetic basis of quantitative traits in maize. BMC genomics 16(1):1078.
9.Hong K, et al. (2015) Transcriptome characterization and expression profiles of the related defense genes in postharvest mango fruit against Colletotrichum gloeosporioides. Gene.
10.Xu W, Dai M, Li F, & Liu A (2014) Genomic imprinting, methylation and parent-of-origin effects in reciprocal hybrid endosperm of castor bean. Nucleic Acids Res 42(11):6987-6998.
11.Qin C, et al. (2014) Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. PNAS 111(14):5135-5140.
12.Lu X, et al. (2014) De novo transcriptome assembly for rudimentary leaves in Litchi chinesis Sonn. and identification of differentially expressed genes in response to reactive oxygen species. BMC genomics 15:805.
13.Ling HQ, et al. (2013) Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496(7443):87-90.
14.Jia J, et al. (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496(7443):91-95.
15.He N, et al. (2013) Draft genome sequence of the mulberry tree Morus notabilis. Nature communications 4:2445.
16.Gao ZY, et al. (2013) Dissecting yield-associated loci in super hybrid rice by resequencing recombinant inbred lines and improving parental genome sequences. PNAS 110(35):14492-14497.
更多基迪奥的原创文章,可继续关注我们网站动态发布,同时关注基迪奥微信~扫一扫添加基迪奥好友~随时随地关注行业动态!